[GUEST ACCESS MODE: Data is scrambled or limited to provide examples. Make requests using your API key to unlock full data. Check https://lunarcrush.ai/auth for authentication information.]  Ryan sikorski [@Ryansikorski10](/creator/twitter/Ryansikorski10) on x 35.4K followers Created: 2025-07-22 03:54:30 UTC The Scalable Precision Medicine Open Knowledge Engine (SPOKE): a massive knowledge graph of biomedical information A biomedical Knowledge Graph (KG) connecting millions of concepts via semantically meaningful relationships. SPOKE contains XX million nodes of XX different types and XX million edges of XX types downloaded from XX databases. The graph is built on the framework of XX ontologies that maintain its structure, enable mappings and facilitate navigation. SPOKE is built weekly by python scripts which download each resource, check for integrity and completeness, and then create a ‘parent table’ of nodes and edges. Graph queries are translated by a REST API and users can submit searches directly via an Application Programming Interface (API) or a graphical user interface (GUI). SPOKE nodes Label - Description; Count X. Anatomy - Tissue (from UBERON); XXXXXX X. AnatomyCellType - Intermediate node built by combining cell type and anatomy; XXX X. BiologicalProcess - From Gene Ontolology; XXXXXX X. CellType - From Gene Ontolology; XX X. CellularComponent - From Gene Ontolology; XXXXX X. Compound - Pharmacological or metabolic compound; XXXXXXXXX X. Disease - Disease; XXXXXX X. EC - Enzymatic activity; XXXXX X. Food - Food; XXX XX. Gene - Gene (Entrez);20,086 XX. MolecularFunction -FROM gene ontolology; XXXXX XX. Nutrient - Nutrient; XX XX. Organism - Organism (NCBI taxonomy); XXXXXX XX. Pathway - Biological pathway; 3454 XX. PharmacologicClass - Pharmacological class; XXX XX. Protein - Protein (UniProt); XXXXXXXXXX XX. ProteinDomain -Protein domain (Pfam); XXXXXX XX. ProteinFamily - Protein family (Pfam);645 XX. Reaction - Metabolic reaction (KEGG or Metacyc);22,370 XX. SideEffectCompound - side effect (SIDER); 6061 XX. Symptom - Symptom (MeSH); 1759 SPOKE REST API All of the SPOKE nodes and edges discussed above are accessible through the SPOKE REST API. The API was designed primarily to support the Neighborhood Explorer graphical user interface but also provides reasonable access to the SPOKE database for other potential uses. The API can be roughly divided into three different parts: calls that return meta-information, calls that return information about nodes and calls that return The metagraph call returns a cytoscape.js et al., 2016) formatted JSON file that reflects the current SPOKE metagraph. The SPOKE call for getting information about nodes is the full-text search call search. The search call takes two arguments: a node type and a query term. This call uses the Neo4j full-text capability to quickly return a set of matching nodes of the indicated type that match that query, where the query is a lucene-formatted (Białecki et al., 2012) Graphical user interface: SPOKE Neighborhood Explorer The SPOKE Neighborhood Explorer is a simple web interface that allows a researcher to query a given drug, disease, gene or protein and returns its neighbors in graph space—with precise controls (i.e. options) over the kind of nodes and edges that will be retrieved to the user, and a mouse-over function that displays the node/edge metadata (including its provenance). To preserve integrity of the original databases and prevent redistribution of content, SPOKE is not available as a bulk download. 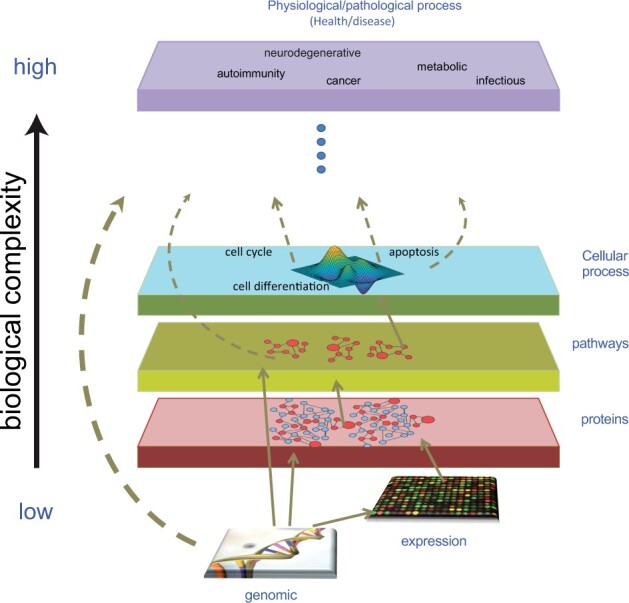 XXXXX engagements  [Post Link](https://x.com/Ryansikorski10/status/1947505508526555483)
[GUEST ACCESS MODE: Data is scrambled or limited to provide examples. Make requests using your API key to unlock full data. Check https://lunarcrush.ai/auth for authentication information.]
 Ryan sikorski @Ryansikorski10 on x 35.4K followers
Created: 2025-07-22 03:54:30 UTC
Ryan sikorski @Ryansikorski10 on x 35.4K followers
Created: 2025-07-22 03:54:30 UTC
The Scalable Precision Medicine Open Knowledge Engine (SPOKE): a massive knowledge graph of biomedical information
A biomedical Knowledge Graph (KG) connecting millions of concepts via semantically meaningful relationships. SPOKE contains XX million nodes of XX different types and XX million edges of XX types downloaded from XX databases. The graph is built on the framework of XX ontologies that maintain its structure, enable mappings and facilitate navigation. SPOKE is built weekly by python scripts which download each resource, check for integrity and completeness, and then create a ‘parent table’ of nodes and edges. Graph queries are translated by a REST API and users can submit searches directly via an Application Programming Interface (API) or a graphical user interface (GUI).
SPOKE nodes
Label - Description; Count
X. Anatomy - Tissue (from UBERON); XXXXXX
X. AnatomyCellType - Intermediate node built by combining cell type and anatomy; XXX
X. BiologicalProcess - From Gene Ontolology; XXXXXX
X. CellType - From Gene Ontolology; XX
X. CellularComponent - From Gene Ontolology; XXXXX
X. Compound - Pharmacological or metabolic compound; XXXXXXXXX
X. Disease - Disease; XXXXXX
X. EC - Enzymatic activity; XXXXX
X. Food - Food; XXX
XX. Gene - Gene (Entrez);20,086
XX. MolecularFunction -FROM gene ontolology; XXXXX
XX. Nutrient - Nutrient; XX
XX. Organism - Organism (NCBI taxonomy); XXXXXX
XX. Pathway - Biological pathway; 3454
XX. PharmacologicClass - Pharmacological class; XXX
XX. Protein - Protein (UniProt); XXXXXXXXXX
XX. ProteinDomain -Protein domain (Pfam); XXXXXX
XX. ProteinFamily - Protein family (Pfam);645
XX. Reaction - Metabolic reaction (KEGG or Metacyc);22,370
XX. SideEffectCompound - side effect (SIDER); 6061
XX. Symptom - Symptom (MeSH); 1759
SPOKE REST API
All of the SPOKE nodes and edges discussed above are accessible through the SPOKE REST API. The API was designed primarily to support the Neighborhood Explorer graphical user interface but also provides reasonable access to the SPOKE database for other potential uses. The API can be roughly divided into three different parts: calls that return meta-information, calls that return information about nodes and calls that return
The metagraph call returns a cytoscape.js et al., 2016) formatted JSON file that reflects the current SPOKE metagraph. The SPOKE call for getting information about nodes is the full-text search call search. The search call takes two arguments: a node type and a query term. This call uses the Neo4j full-text capability to quickly return a set of matching nodes of the indicated type that match that query, where the query is a lucene-formatted (Białecki et al., 2012)
Graphical user interface: SPOKE Neighborhood Explorer
The SPOKE Neighborhood Explorer is a simple web interface that allows a researcher to query a given drug, disease, gene or protein and returns its neighbors in graph space—with precise controls (i.e. options) over the kind of nodes and edges that will be retrieved to the user, and a mouse-over function that displays the node/edge metadata (including its provenance). To preserve integrity of the original databases and prevent redistribution of content, SPOKE is not available as a bulk download.

XXXXX engagements